Starting example: square¶
This small example will you demonstrate the basic methodology how our package works. We use a very simple model represented by only one function that squares the input data and we create a project to investigate trigonmetric functions (sine, cosine, etc.).
So let’s define our model function
"""A small example for a computational model
"""
def compute(x):
return x ** 2
and save it in a file called 'calculate.py' file. This function runs
only in python and does not know anything about where the input is from and
where it is saved.
We start from the pure ModelOrganizer to manage our model named
square
#!/usr/bin/env python
from model_organization import ModelOrganizer
class SquareModelOrganizer(ModelOrganizer):
name = 'square'
if __name__ == '__main__':
SquareModelOrganizer.main()
and save it in the file called 'square.py'. If we run our model from the
command line, (for technical reasons we run it here from inside IPython), we
see already several preconfigured commands
In [1]: !./square.py -h
usage: square [-h] [-id str] [-l] [-n] [-v] [-vl str or int] [-nm] [-E]
{setup,init,set-value,get-value,del-value,info,unarchive,configure,archive,remove}
...
The main function for parsing global arguments
positional arguments:
{setup,init,set-value,get-value,del-value,info,unarchive,configure,archive,remove}
setup Perform the initial setup for the project
init Initialize a new experiment
set-value Set a value in the configuration
get-value Get one or more values in the configuration
del-value Delete a value in the configuration
info Print information on the experiments
unarchive Extract archived experiments
configure Configure the project and experiments
archive Archive one or more experiments or a project instance
remove Delete an existing experiment and/or projectname
optional arguments:
-h, --help show this help message and exit
-id str, --experiment str
experiment: str
The id of the experiment to use. If the `init` argument is called, the `new` argument is automatically set. Otherwise, if not specified differently, the last created experiment is used.
-l, --last If True, the last experiment is used
-n, --new If True, a new experiment is created
-v, --verbose Increase the verbosity level to DEBUG. See also `verbosity_level`
for a more specific determination of the verbosity
-vl str or int, --verbosity-level str or int
The verbosity level to use. Either one of ``'DEBUG', 'INFO',
'WARNING', 'ERROR'`` or the corresponding integer (see pythons
logging module)
-nm, --no-modification
If True/set, no modifications in the configuration files will be
done
-E, --match If True/set, interprete `experiment` as a regular expression
(regex) und use the matching experiment
So to setup our new project, we use the setup() method
In [2]: !./square.py setup . -p trigo
INFO:square.trigo:Initializing project trigo
And we initialize one experiment for the sine, one for the cosine, and one for the tangent functions
In [3]: !./square.py -id sine init -d "Squared Sine"
INFO:square.sine:Initializing experiment sine of project trigo
In [4]: !./square.py -id cosine init -d "Squared Cosine"
����������������������������������������������������������������INFO:square.cosine:Initializing experiment cosine of project trigo
In [5]: !./square.py -id tangent init -d "Squared Tangent"
������������������������������������������������������������������������������������������������������������������������������������INFO:square.tangent:Initializing experiment tangent of project trigo
Now of course, we, need the input data, so we enhance our
ModelOrganizer subclass with a preprocess method
#!/usr/bin/env python
import os.path as osp
import numpy as np
from model_organization import ModelOrganizer
class SquareModelOrganizer(ModelOrganizer):
name = 'square'
commands = ModelOrganizer.commands[:]
# insert the new run command to the other commands, right before the
# archiving
commands.insert(commands.index('archive'), 'preproc')
def preproc(self, which='sin', **kwargs):
"""
Create preprocessing data
Parameters
----------
which: str
The name of the numpy function to apply
``**kwargs``
Any other parameter for the
:meth:`model_organization.ModelOrganizer.app_main` method
"""
self.app_main(**kwargs)
config = self.exp_config
config['infile'] = infile = osp.join(config['expdir'], 'input.dat')
func = getattr(np, which) # np.sin, np.cos or np.tan
data = func(np.linspace(-np.pi, np.pi))
self.logger.info('Saving input data to %s', infile)
np.savetxt(infile, data)
def _modify_preproc(self, parser):
"""Modify the parser for the preprocessing command"""
parser.update_arg('which', short='w', choices=['sin', 'cos', 'tan'])
if __name__ == '__main__':
SquareModelOrganizer.main()
The code should be more or less self explanatory. We start with the
ModelOrganizer.app_main() method which chooses the right experiment from
the given id parameter. The we use the numpy.sin, numpy.cos and
numpy.tan functions from the numpy module and calculate the data from
\(-\pi\) to \(\pi\).
Finally we store it to an input file inside the experiment directory.
Hint
If you want to see the configuration values in the experiment config, use
the info() and the
get-value commands
In [6]: !./square.py info
id: tangent
description: Squared Tangent
project: trigo
expdir: /home/docs/checkouts/readthedocs.org/user_builds/model-organization/checkouts/latest/docs/trigo/experiments/tangent
timestamps:
init: '2019-05-23 22:32:31.351991'
setup: '2019-05-23 22:32:29.343508'
In [7]: !./square.py get-value expdir
����������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������/home/docs/checkouts/readthedocs.org/user_builds/model-organization/checkouts/latest/docs/trigo/experiments/tangent
In the _modify_preproc section, we update argument of the --which
argument to include a shorter -w command and some choices (see the
funcargparse.FuncArgParser class for more information). Note the style
of the documentation of the preproc method. We follow the
numpy documentation guidelines such that the
funcargparse.FuncArgParser can extract the docstring. Our new command
now has been translated to a command line argument:
In [8]: !./square.py preproc -h
��������������������usage: square preproc [-h] [-w str]
Create preprocessing data
optional arguments:
-h, --help show this help message and exit
-w str, --which str The name of the numpy function to apply
and we can use it to create our input data
In [9]: !./square.py -id sine preproc
INFO:square.sine:Saving input data to /home/docs/checkouts/readthedocs.org/user_builds/model-organization/checkouts/latest/docs/trigo/experiments/sine/input.dat
In [10]: !./square.py -id cosine preproc -w cos
������������������������������������������������������������������������������������������������������������������������������������������������������������������INFO:square.cosine:Saving input data to /home/docs/checkouts/readthedocs.org/user_builds/model-organization/checkouts/latest/docs/trigo/experiments/cosine/input.dat
# by default, the last created experiment (tangent) is used so the -id
# argument is not necessary
In [11]: !./square.py preproc -w tan
����������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������INFO:square.tangent:Saving input data to /home/docs/checkouts/readthedocs.org/user_builds/model-organization/checkouts/latest/docs/trigo/experiments/tangent/input.dat
Finally, we include a run command that uses our input data of the given
experiment and for our model
def run(self, **kwargs):
"""
Run the model
Parameters
----------
``**kwargs``
Any other parameter for the
:meth:`model_organization.ModelOrganizer.app_main` method
"""
from calculate import compute
self.app_main(**kwargs)
# get the default output name
output = osp.join(self.exp_config['expdir'], 'output.dat')
# save the paths in the configuration
self.exp_config['output'] = output
# run the model
data = np.loadtxt(self.exp_config['infile'])
out = compute(data)
# save the output
self.logger.info('Saving output data to %s', osp.relpath(output))
np.savetxt(output, out)
# store some additional information in the configuration of the
# experiment
self.exp_config['mean'] = mean = float(out.mean())
self.exp_config['std'] = std = float(out.std())
self.logger.debug('Mean: %s, Standard deviation: %s', mean, std)
and we run it
In [12]: !./square.py -v -id sine run
���������������������INFO:square.sine:Saving output data to trigo/experiments/sine/output.dat
DEBUG:square.sine:Mean: 0.49000000000000016, Standard deviation: 0.35693136595149494
In [13]: !./square.py -v -id cosine run
�������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������INFO:square.cosine:Saving output data to trigo/experiments/cosine/output.dat
DEBUG:square.cosine:Mean: 0.51, Standard deviation: 0.35693136595149494
In [14]: !./square.py -v run
��������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������INFO:square.tangent:Saving output data to trigo/experiments/tangent/output.dat
DEBUG:square.tangent:Mean: 47.04000000000017, Standard deviation: 190.14783301421124
Now, finally, let’s create a postproc command that visualizes our data
def postproc(self, close=True, **kwargs):
"""
Postprocess and visualize the data
Parameters
----------
close: bool
Close the figure at the end
``**kwargs``
Any other parameter for the
:meth:`model_organization.ModelOrganizer.app_main` method
"""
import matplotlib.pyplot as plt
import seaborn as sns # for nice plot styles
self.app_main(**kwargs)
# ---- load the data
indata = np.loadtxt(self.exp_config['infile'])
outdata = np.loadtxt(self.exp_config['output'])
x_data = np.linspace(-np.pi, np.pi)
# ---- make the plot
fig = plt.figure()
# plot input data
plt.plot(x_data, indata, label='input')
# plot output data
plt.plot(x_data, outdata, label='squared')
# draw a legend
plt.legend()
# use the description of the experiment as title
plt.title(self.exp_config.get('description'))
# ---- save the plot
self.exp_config['plot'] = ofile = osp.join(self.exp_config['expdir'],
'plot.png')
self.logger.info('Saving plot to %s', osp.relpath(ofile))
fig.savefig(ofile)
if close:
plt.close(fig)
In [15]: !./square.py -v -id sine postproc
INFO:square.sine:Saving plot to trigo/experiments/sine/plot.png
In [16]: !./square.py -v -id cosine postproc
�����������������������������������������������������������������INFO:square.cosine:Saving plot to trigo/experiments/cosine/plot.png
In [17]: !./square.py -v postproc
��������������������������������������������������������������������������������������������������������������������������������������INFO:square.tangent:Saving plot to trigo/experiments/tangent/plot.png
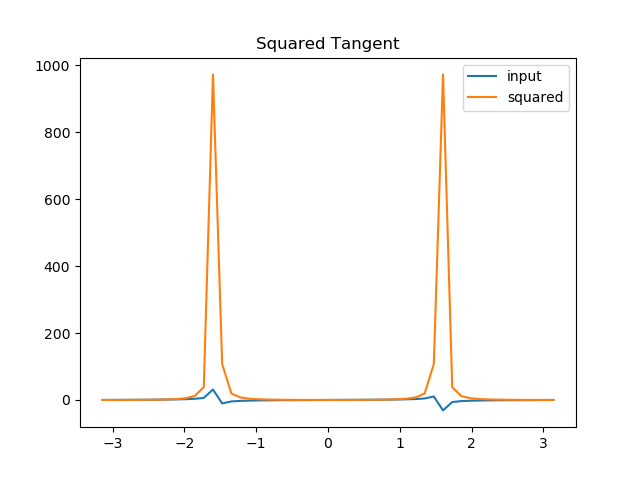
Finally, we can archive our project to save disc space
In [18]: !./square.py archive -p trigo -rm
INFO:square:Archiving to /home/docs/checkouts/readthedocs.org/user_builds/model-organization/checkouts/latest/docs/trigo.tar
Hint
You can also do everything from above in one line by chaining the subparser commands.
In [19]: !./square.py -v -id sine setup . -p trigo init -d "Squared Sine" preproc run postproc archive -p trigo -rm
INFO:square.sine:Initializing project trigo
DEBUG:square.sine: Creating root directory /home/docs/checkouts/readthedocs.org/user_builds/model-organization/checkouts/latest/docs/trigo
INFO:square.sine:Initializing experiment sine of project trigo
DEBUG:square.sine: Creating experiment directory /home/docs/checkouts/readthedocs.org/user_builds/model-organization/checkouts/latest/docs/trigo/experiments/sine
INFO:square.sine:Saving input data to trigo/experiments/sine/input.dat
INFO:square.sine:Saving output data to trigo/experiments/sine/output.dat
DEBUG:square.sine:Mean: 0.49000000000000016, Standard deviation: 0.35693136595149494
INFO:square.sine:Saving plot to trigo/experiments/sine/plot.png
INFO:square.sine:Archiving to /home/docs/checkouts/readthedocs.org/user_builds/model-organization/checkouts/latest/docs/trigo.tar
DEBUG:square.sine:Adding /home/docs/checkouts/readthedocs.org/user_builds/model-organization/checkouts/latest/docs/trigo/experiments/sine
DEBUG:square.sine:Store sine experiment config to /home/docs/checkouts/readthedocs.org/user_builds/model-organization/checkouts/latest/docs/trigo/.project/sine.yml
DEBUG:square.sine:Store trigo project config to /home/docs/checkouts/readthedocs.org/user_builds/model-organization/checkouts/latest/docs/trigo/.project/.project.yml
DEBUG:square.sine:Add /home/docs/checkouts/readthedocs.org/user_builds/model-organization/checkouts/latest/docs/trigo/.project to archive
DEBUG:square.sine:Removing /home/docs/checkouts/readthedocs.org/user_builds/model-organization/checkouts/latest/docs/trigo/experiments/sine
DEBUG:square.sine:Removing /home/docs/checkouts/readthedocs.org/user_builds/model-organization/checkouts/latest/docs/trigo